I Am Healthcare Transparency and So Can You
If you're like me you've spent a lot of the last two months thinking about how to parse huge JSON files. That's because some of the most valuable data in the world of healthcare is buried in them.
These files are big (sometimes 100GB+) and annoying enough that a cottage industry of businesses has sprung up around them: they manage the data munging, storage, and retrieval, and you shell out for the convenience. And getting started on your own really can be a headache.
Free data isn't free
That convenience will cost you at least thousands of dollars a month, at most tens of thousands. What if you wanted to do this on a budget? Well by streaming them and saving just what you need, you can be on your merry way and put those thousands of dollars towards something useful, like your deductibles.
One of the reasons paying for the data outright is so expensive is because there's so much of it -- probably around a petabyte is published monthly.
Junk DNA
But similar to the human genome, most of it is either useless or has some other agenda. You see things like opthamalogists's rates for C-sections, or physical therapists's rates for brain surgery. Rates that are purely an artifact of how contracts are negotiated in healthcare but would never (and could never, legally speaking) be on an actual bill.
So you absolutely cannot and should not try to get every code from every provider. It doesn't make any sense and it's a waste of time. Instead, pick which billing codes you're interested in and find the providers that are likely to offer that service. I'm gonna show you how to do that.
Open question: is there a way to link an NPI number (provider) to the kinds of procedures they can legally perform? If so, we could cut down the size of the data by as much as 20x. If you have a possible answer to this question, write me at alec@dolthub.com.
Okay so how do I do that
You're ready to save the world and make sense of the biggest trove of healthcare data ever -- you just need to know how. I'm gonna walk you through the basic steps and hopefully by the end of this you too will feel empowered to expose the insanity of the US healthcare system. If you don't want to know how any of this works, you can skip to the shell script at the bottom and just run it.
Some have written similar parsers in faster languages than python. In my experience the time to flattening is all download-bandwidth limited so there's no point. Just do it the easiest way.
1. Unzip and stream
We're going to need to process these files as they come in because they're almost always too big to read into memory.
Two ingredients go into making this work. The first is a magical combination of requests and gzip, the second is the JSON-streaming package ijson.
First, we stream the zipped MRFs using this magical two-liner:
with requests.get(url, stream = True) as r:
f = gzip.GzipFile(fileobj = r.raw)
# stream = True and fileobj = r.raw go together.
# see: https://requests.readthedocs.io/en/latest/api/The unzipped file f looks something like (for example):
{
"reporting_entity_name": "United HealthCare Services, Inc.",
"reporting_entity_type": "Third-Party Administrator",
"last_updated_on": "2022-11-01",
"version": "1.0.0",
...2. Whip out ijson
Let's peek at what ijson gives us for free using the parse method. If we run:
parser = ijson.parse(f, use_float = True)
# use_float = True makes sure that numbers like 10.1 get parsed as floats and not decimals
for row in parser:
print(row)you get:
('', 'start_map', None)
('', 'map_key', 'reporting_entity_name')
('reporting_entity_name', 'string', 'United HealthCare Services, Inc.')
('', 'map_key', 'reporting_entity_type')
('reporting_entity_type', 'string', 'Third-Party Administrator')
('', 'map_key', 'last_updated_on')
('last_updated_on', 'string', '2022-11-01')
('', 'map_key', 'version')
('version', 'string', '1.0.0')
...So each "leaf" in the parsed "tree" gets its own tuple representing its value and its location in the file. The 'start_map' and 'map_key' stuff is ijson's way of letting the ObjectBuilder know where it is in the file.
Slight detour: ijson.ObjectBuilder
One of the coolest parts about ijson API is the ObjectBuilder. I definitely recommend checking out the source code because the ObjectBuilder itself is a pretty clever little snippet of code, and worthy of its own discussion, probably.
The ObjectBuilder API pretty much works like this: you create a builder, then every time you get an event in the parser, you add it to the builder through the builder.event method.
with requests.get(url, stream = True) as r:
f = gzip.GzipFile(fileobj = r.raw)
parser = ijson.parse(f, use_float=True)
builder = ijson.ObjectBuilder()
for prefix, event, value in parser:
# ('version', 'string', '1.0.0'), etc
builder.event(event, value)
This turns the entire file into a single python dictionary. The above snippet is the same as just downloading the file and reading the JSON into memory as a dictionary.
So now with those basics in hand (reading the file, building objects, etc.) we can check out the nitty-gritty of handling the individual parts of the file processing.
Relational tables
To make these prices workable we'll need to flatten them out of the JSON format and into something easier. I chose CSV for simplicity. We'll make one CSV file for each conceptual block:
- the root data metadata
- the billing codes
- the prices
The solution to keying the tables: hashes
We'll need keys to link these tables, and the keys have to have the following properties.
- If two computers flatten the same files they must produce the same keys. This rules out UUIDs
- If two files are flattened by the same computer, they must produce different keys. This rules out ints
So I went with hashes. Here's how it keys each row, with comments as examples:
def hashdict(data_dict):
# data_dict = {'b': 2, 'a': 1}
sorted_tups = sorted(data_dict.items())
# [('a', 1), ('b', 2)]
dict_as_bytes = json.dumps(sorted_tups).encode("utf-8")
# b'[["a", 1], ["b", 2]]'
dict_hash = hashlib.sha256(dict_as_bytes).hexdigest()[:16]
# 91cea03dbc87dd3d
return dict_hashFlattening and writing
Let's start with the easiest case, building the dictionary for the root data (the file metadata that's at the top of every JSON file). We pass the parser to a build root function that will stop once it hits the 'start_array' event (which tells us we're no longer in the metadata):
def build_root(parser):
builder = ijson.ObjectBuilder()
for prefix, event, value in parser:
if event == "start_array":
# once we hit "start_array",
# we've passed the root metadata
# return the dict as builder.value
# and the last item in the tree (prefix, event, value)
return builder.value, (prefix, event, value)
builder.event(event, value)Then we take those values and key them with our hashdict function:
root_vals, row = build_root(parser)
# get a hash of the data and
# store it in the dictionary
root_hash_id = hashdict(root_vals)
root_vals["root_hash_id"] = root_hash_idTo write this to file, we pass this dictionary to a special function which turns it into rows (and adds all the hash_ids necessary), then another function which writes it to file:
So that's the basics. But how do we filter as we go?
Filtering as we go
We build in-network objects the same way, except we add a little trick for skipping over the parts we don't need to save (irrelevant parts elided):
def build_innetwork(init_row, parser, code_list=None, npi_list=None, provref_idx=None):
builder = ijson.ObjectBuilder()
builder.event(event, value)
for nprefix, event, value in parser:
...
# return early if we hit codes that aren't
# in code_list
elif nprefix.endswith("negotiated_rates") and event == "start_array":
if code_list:
billing_code_type = builder.value["billing_code_type"]
billing_code = str(builder.value["billing_code"])
if (billing_code_type, billing_code) not in code_list:
return None, (nprefix, event, value)
...
# skip over any npi_numbers that aren't
# in the npi_list
elif nprefix.endswith("npi.item"):
if npi_list and value not in npi_list:
continue
...You can then take this dict of in-network data, turn it into rows with the aforementioned special function:
def innetwork_to_rows(obj, root_hash_id):
rows = []
in_network_vals = {
"negotiation_arrangement": obj["negotiation_arrangement"],
"name": obj["name"],
"billing_code_type": obj["billing_code_type"],
"billing_code_type_version": obj["billing_code_type_version"],
"billing_code": obj["billing_code"],
"description": obj["description"],
"root_hash_id": root_hash_id,
}
in_network_hash_id = hashdict(in_network_vals)
in_network_vals["in_network_hash_id"] = in_network_hash_id
rows.append(("in_network", in_network_vals))
...
# continue for the other tables
# (omitted for brevity)
return rowsand write them to file.
The rest of the program works the same way but with some extra details.
- It takes the provider references and filters the unwanted NPI numbers out
- If swaps the provider references for the actual provider information (increasing space usage, but eliminating a table)
- It gets provider references that exist in remote files (yea, this is actually a thing that they can do)
The functions in core.py implement all of this out-of-the-box. Feel free to read through some of the code to see how it works.
An analysis of dialysis: an example that you can run right now
Go ahead and clone this repo and run the dialysis.sh script in the /python/processors/dialysis_example folder, and read the rest of this while you wait for it to finish.
Getting the NPI numbers you need
So let's talk about how to use this program to actually get something you want. Say we want to take a look at dialysis: CPT codes 90935, 90937, and 90940.
Not everyone offers dialysis: most hospitals (probably) do, as do dialysis clinics. We conveniently have hospital NPI numbers on hand already. And we'll throw in all the known dialysis centers for good measure by pulling them out of the CMS NPI registry:
import polars as pl
(pl
.scan_csv('npidata_pfile_20050523-20220911.csv')
.filter(pl.col('Healthcare Provider Taxonomy Code_1') == '2472R0900X') # Dialysis clinics
.filter(pl.col('Entity Type Code') == '2') # only organizations, not individuals
.select(['NPI'])
.collect()).to_csv('dialysis.csv')Now let's go ahead and run an example. Check out this script which has everything you need already built-in.
python process_dialysis.py -u https://uhc-tic-mrf.azureedge.net/public-mrf/2022-09-01/2022-09-01_UnitedHealthcare-of-Texas--Inc-_Insurer_HML-75_ED_in-network-rates.json.gz -o dialysis_data --npi dialysis_npis.csvAll you need now is your list of URLs.
Getting the URLs
Conveniently you can run one of the url downloaders to get a list of URLs to scrape. It'll put those URLs in a SQLite database for you to page through.
I went ahead and pulled a sample of 100 and sent them to urls.txt.
df = pl.read_csv('uhc_filtered_urls.csv')['url'].sample(100)
with open('urls.txt', 'w') as f:
for row in df:
f.write(row+'\n')Running this script on lots of URLs at once
One way to run these downloaders in parallel is to run them via screen. Take your urls.txt, a simple list of URLs, then create a shell script like this one:
#!/bin/bash
for URL in $(cat urls.txt)
do
screen -dm python3 process_dialysis.py -o dialysis --npi dialysis_npis.csv -u $URL
doneThis'll output the data into the folder dialysis, which we can take a peek at.
A brief analysis of dialysis
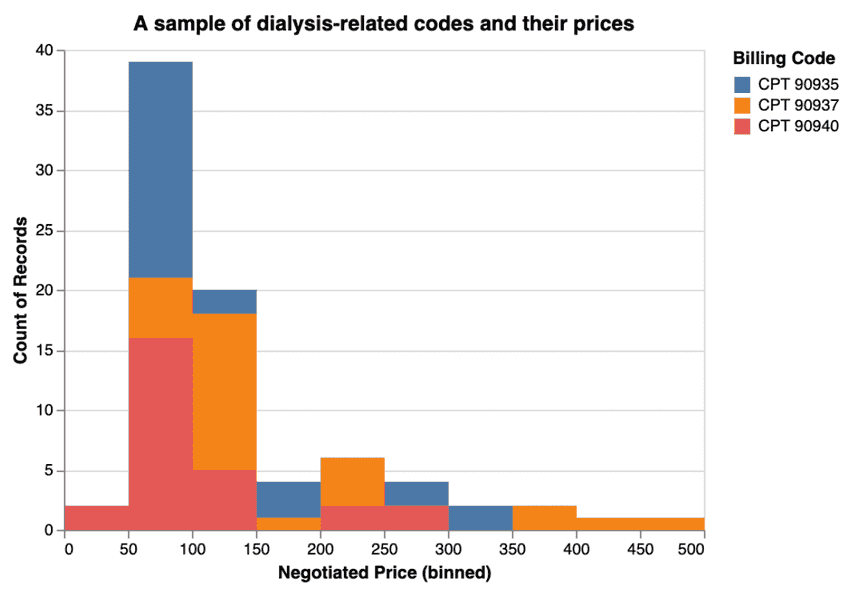
You're ready to make your first chart. Here's an example of what you can do with Altair and a Jupyter notebook. You can take those CPT codes above (two of which represent hemodialysis and one (90940) which is a related billing code) and plot them on a chart to see how much they get billed for.
Dialysis prices fresh out of the oven: it's likely no one has ever seen these prices before.
Now the real question, and where you come in, is to interpret this data. You can pick a topic like I did for C-sections or write about whatever case suits your interest -- business, journalistic, or otherwise.
Does it make sense that for one round of dialysis, some facilities charge $50, while others charge ten times that much? Can you find correlations? Connections?
I'm starting to come around to the way of thinking that this can't be the project of one person or even one team, but that we should all be picking away at this data together to make sense of it.
Next steps
Have thoughts or want help getting started? Join our #datasets Discord channel to talk more about this. We're looking for people who can help us massage this data into a more workable form.
If you want to join one of our healthcare-transparency-related data bounties, we just started one in collaboration with the nonprofit OneFact to collect hospital chargemaster URLs. Hop on our #data-bounties channel to learn more.